GenAI – What Is Multi Hop RAG Architecture ?
Scenario:
- How To Design The Multi Hop RAG System ?
Answer:


Why We Need Multi Hop RAG ?

How Does Multi Hop RAG Works ?





How Does Multi Hop RAG Connects To Different Document Sources ?




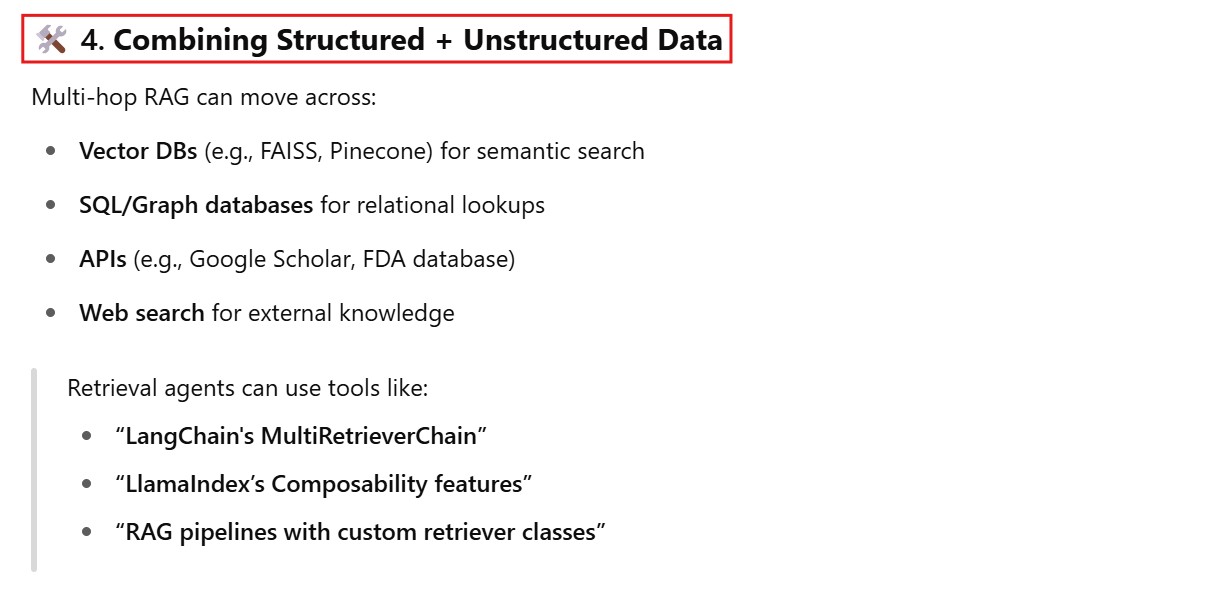

How to configure each retrieval hop to query a different vector database, search engine, or API, depending on the nature of the sub-question ?





from langchain.chains import RetrievalQA
from langchain.vectorstores import FAISS
from langchain.agents import initialize_agent, Tool
# Define different retrievers
medical_vector_db = FAISS.load_local("path_to_medical_vector_store")
search_engine = ElasticSearchRetriever(index_name="company_data")
api_fda = APIRetriever("https://api.fda.gov")
general_search = GoogleCustomSearchAPI()
# Define the retrieval chain that chooses the retriever dynamically
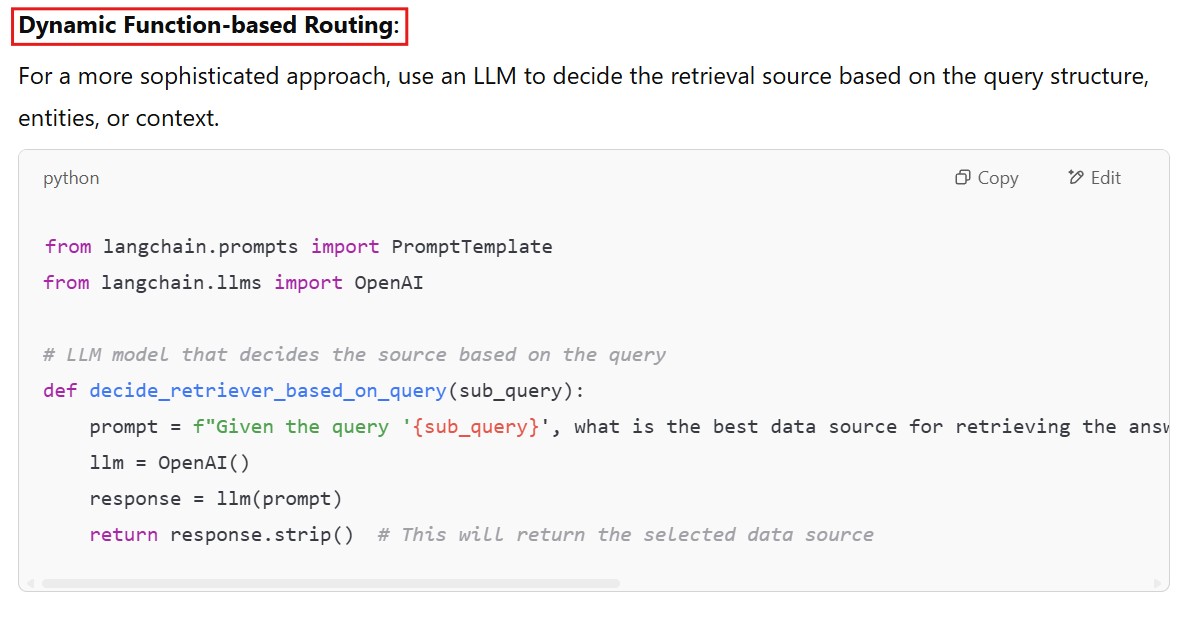
def get_answer_from_retriever(sub_query):
retriever_choice = decide_retriever_based_on_query(sub_query) # This will return the source
if retriever_choice == 'medical_vector_db':
retriever = medical_vector_db.as_retriever()
elif retriever_choice == 'search_engine':
retriever = search_engine
elif retriever_choice == 'api_fda':
retriever = api_fda
else:
retriever = general_search
# Create the retrieval chain
qa_chain = RetrievalQA(retriever=retriever, llm=OpenAI())
# Get the answer from the selected retriever
return qa_chain.run(sub_query)

# Hop 1: Retrieve medical symptoms for Long COVID
hop_1_query = "What are the symptoms of long COVID?"
hop_1_output = get_answer_from_retriever(hop_1_query)
# Hop 2: Use the results of Hop 1 to search for FDA-approved treatments
hop_2_query = f"What FDA-approved treatments address {hop_1_output}?"
hop_2_output = get_answer_from_retriever(hop_2_query)
# Final output
print(f"Final Answer: {hop_2_output}")
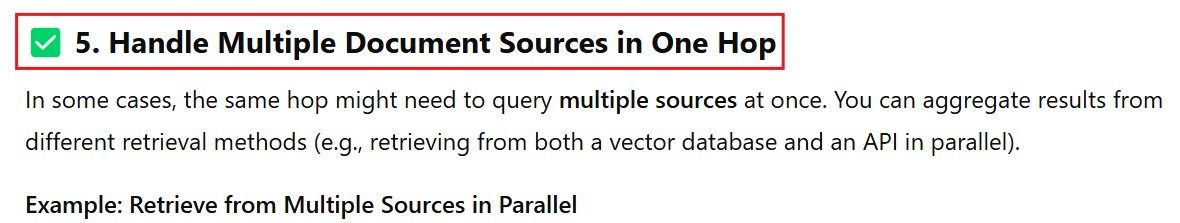
from langchain.chains import ParallelChain
def parallel_retrieval(sub_query):
retrievers = [
medical_vector_db.as_retriever(),
search_engine,
api_fda
]
# Perform parallel retrieval
parallel_chain = ParallelChain(retrievers)
results = parallel_chain.run(sub_query)
return results


Example Of Multi Hop RAG Process .



from langchain.llms import OpenAI
from langchain.vectorstores import FAISS
from langchain.agents import initialize_agent, Tool
from langchain.chains import RetrievalQA
from langchain.prompts import PromptTemplate
# Set up different data sources (retrievers)
medical_vector_db = FAISS.load_local("path_to_medical_vector_store") # Example vector DB (e.g., FAISS)
search_engine = ElasticSearchRetriever(index_name="treatments_data") # Custom search engine
api_fda = APIRetriever("https://api.fda.gov") # FDA Database API
# Function to decide which data source to query based on the sub-query
def choose_retriever(sub_query):
if "symptoms" in sub_query.lower():
return medical_vector_db.as_retriever() # Use vector database for symptoms
elif "FDA" in sub_query.lower():
return api_fda # Use FDA API for treatments
else:
return search_engine # Default search engine for other queries
# Function to perform multi-hop query
def get_answer_from_retriever(sub_query):
retriever_choice = choose_retriever(sub_query) # Decide the retriever based on the sub-query
qa_chain = RetrievalQA(retriever=retriever_choice, llm=OpenAI()) # Use an LLM to answer with the retrieved data
return qa_chain.run(sub_query) # Run the chain to get the answer

# Hop 1: Find the symptoms of long COVID from the medical vector database
hop_1_query = "What are the symptoms of long COVID?"
hop_1_output = get_answer_from_retriever(hop_1_query)
# Hop 2: Use the symptoms from Hop 1 to query the FDA API for treatments
hop_2_query = f"What FDA-approved treatments are available for {hop_1_output}?"
hop_2_output = get_answer_from_retriever(hop_2_query)
# Final answer
print(f"FDA-approved treatments for long COVID symptoms: {hop_2_output}")




